Product Name: Host Cell Residual DNA Sample Preprocessing Kit (Magnetic Bead Method)
The residual DNA of host cells in biological products has many risks such as tumorigenicity and infectivity, so the accurate quantitative detection of trace amounts of residual DNA is particularly important. Pretreatment is the process of extracting and purifying trace amounts of DNA in biological products from complex sample matrices. An effective and stable pretreatment method is the basis for ensuring accurate detection of residual DNA detection and other rapid nucleic acid detection methods.
BlueKit Host Cell Residual DNA Sample Preprocessing Kit can meet both manualextraction and machine extraction methods. Manual extraction is accurate and sensitive, and it iefficient and convenient to use with a fully automatic nucleic acid extractor.
Ready-to-use standards
Wide Linear range
One-step design
Calibrated with international NIBSC/WHO (90/636) standards
Validated kit performance
Proven 10+ IND application experience
Developed with products pecificity
Short detection time
Error-proofdesign
1. Sample Prep: Dilution/pH adjustment (pH 6.0–8.0)
2. Lysis: Proteinase K + 65°C incubation
3. DNA Binding: Magnetic beads + isopropanol
4. Wash/Ethanol Removal
5. Elution: 70°C, high recovery.
| Category | Specifications |
|---|---|
| Intended Use | Pretreatment of biological samples (e.g., CHO, E. coli, Vero, Human cells, plasmids) for accurate extraction of trace host cell residual DNA. Compatible with downstream qPCR detection kits. For research use only. |
| Key Features | - High-sensitivity magnetic bead-based DNA extraction - Compatible with diverse sample matrices (buffers, dry powders) - Includes glycogen & yeast tRNA to minimize loss of low-concentration DNA - Scalable for high-throughput workflows. |
| Kit Components | - Lysis Buffer (9 mL) - Wash Buffer (30 mL, ethanol required) - Eluent (12 mL) - Magnetic Beads (2 × 1 mL) - Proteinase K (1 mL) - Glycogen (2 × 800 μL, -20°C) - Yeast tRNA (50 μL, -20°C). |
| Storage & Shelf Life | 12 months at 2–8°C (except glycogen/tRNA: -20°C or below). |
| Equipment Needed | - Vortex mixer, magnetic stand, centrifuge - 80% ethanol, isopropanol, PBS buffer - Low-attachment tubes, sterile tips. |
| Workflow | 1. Sample Prep: Dilution/pH adjustment (pH 6.0–8.0) 2. Lysis: Proteinase K + 65°C incubation 3. DNA Binding: Magnetic beads + isopropanol 4. Wash/Ethanol Removal 5. Elution: 70°C, high recovery. |
| QC & Yield | - Triplicate extraction recommended - Compatible with SDS-PAGE/Western Blot (if tagged) - Deliverables: Purified DNA in elution buffer, ready for qPCR. |
| Turnaround Time | - Standard: 1–2 days (per batch) - High-throughput: Customizable. |
| Advantages vs. Competitors | - Higher sensitivity: Glycogen/tRNA reduces DNA loss - Broad compatibility: Works with CHO, E. coli, plasmids, etc. - Optimized protocols: Includes spiking controls and neutral pH guidance. |
Purpose: Ensure sensitive and accurate extraction of trace host cell DNA (e.g., CHO, E. coli, Vero, human, plasmid) from complex biological matrices.
Feature:Proprietary magnetic beads with high DNA-binding capacity (>95% recovery) and minimal inhibitor carryover.
Integrated glycogen + yeast tRNA binding buffer to enhance DNA yield for low-concentration samples.
Application: Compatible with downstream qPCR/ddPCR detection (e.g., Hillgene’s DNA detection kits).
Technical Effort:
Validated for diverse samples: upstream intermediates, dry powders (10–100 mg/mL), and neutral-pH-adjusted solutions.
Parallel processing (triplicate recommended) ensures reproducibility.
Purpose: Minimize cross-contamination and maximize operational consistency.
Feature:Pre-optimized lysis + proteinase K digestion (65°C/15 min) for complete cell disruption.
Ethanol-adjusted wash buffers and low-retention tips reduce sample loss.
Application: Ideal for GMP-compliant labs requiring trace DNA analysis in biologics.
Technical Effort:
Includes magnetic stand protocols for rapid bead separation (<5 min/sample).
Room-temperature drying (3–5 min) or blast drying (2 min) prevents over-drying artifacts.
Purpose: Adapt to variable sample volumes and high-throughput needs.
Feature:Linear dynamic range: Handles 100–1000× diluted high-DNA samples (e.g., upstream intermediates).
Spike-in recovery validation (2–10× sample DNA concentration) for QC compliance.
Application: Supports biologics R&D, biosafety testing, and process validation.
Technical Effort:
Kit components stable for 12 months at 2–8°C/-20°C.
Compatible with automated liquid handlers (e.g., KingFisher systems) for scaled processing.
Background
Residual host cell DNA (HCD) in biologics poses risks for immunogenicity and regulatory non-compliance. CHO cells are widely used in bioproduction, but their complex lysates challenge DNA extraction due to high protein/lipid content.
Key Challenge
Conventional column-based kits suffer from low recovery (<50%) for trace DNA (<10 pg/μL) in viscous CHO samples, leading to inconsistent qPCR results.
Optimization
Hillgene’s magnetic bead-based pretreatment kit leverages:
Glycogen/tRNA carrier system to enhance micro-DNA binding.
Lysis buffer + Proteinase K for complete chromatin dissociation.
Ethanol-free elution to minimize inhibitor carryover for downstream qPCR.
Result
Recovery rate: 92.5% (spiked DNA: 5–100 pg/μL).
Purity: A260/A280 = 1.8±0.1, compatible with CHO-specific qPCR kits.
Throughput: 48 samples processed in <1 hour.
Background
Plasmid DNA purification requires stringent HCD clearance (<1 ng/dose). E. coli endotoxins and polysaccharides interfere with DNA extraction.
Key Challenge
Commercial kits fail to remove enzymatic inhibitors, causing false negatives in qPCR for low-abundance HCD (<0.1%).
Optimization
Hillgene’s kit features:
Isopropanol-enhanced binding for fragments <100 bp.
Dual wash buffers to eliminate endotoxins/polysaccharides.
Magnetic bead uniformity (CV <5% lot-to-lot) for reproducible yields.
Result
LOD: 0.05 pg/μL (linear range: 0.1–500 pg/μL).
Inhibitor removal: >99% reduction in LPS contamination.
Regulatory compliance: Meets USP <1130> and EP 2.6.34 guidelines.
Background
Complex biologics (e.g., vaccines, viral vectors) contain HCD from diverse hosts (Vero, human cell lines, SV40).
Key Challenge
Matrix interference (e.g., serum proteins, PEG) reduces DNA recovery variability across sample types.
Optimization
Kit advantages:
Universal lysis buffer for CHO/Vero/human/SV40 samples.
pH adaptability (6.0–8.0) for denatured or neutralized samples.
Pre-spiked glycogen to stabilize nano-DNA during extraction.
Result
Consistency: 85–95% recovery across 10+ biologics (serum, cell pellets, lyophilized powders).
Interoperability: Seamless integration with Hillgene’s qPCR kits (e.g., SV40LTA detection).
Scalability: Validated for 1 mL to 200 μL sample volumes.
| Specification | 100 Reactions |
Detection sensitivity | 0.03pg/μL |
Recovery rate | 70%~130% |
| Validity period | 12 months |
| Storage conditions | RT & 4℃&-20℃ |
| LOQ | NA |
| LOD | NA |
| IND Filing | OK |
| FDA Filing | NA |
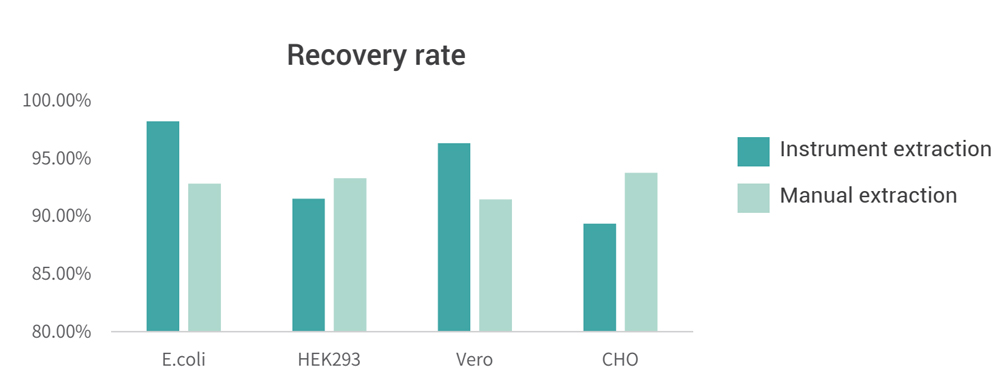
 Manual extraction and instrument extraction were performed on DNA samples of different hosttypes, and the final sample recovery rates were 70% to 130%, which were better than the 50% to 150% required by the Pharmacopoeia.
Manual extraction and instrument extraction were performed on DNA samples of different hosttypes, and the final sample recovery rates were 70% to 130%, which were better than the 50% to 150% required by the Pharmacopoeia.
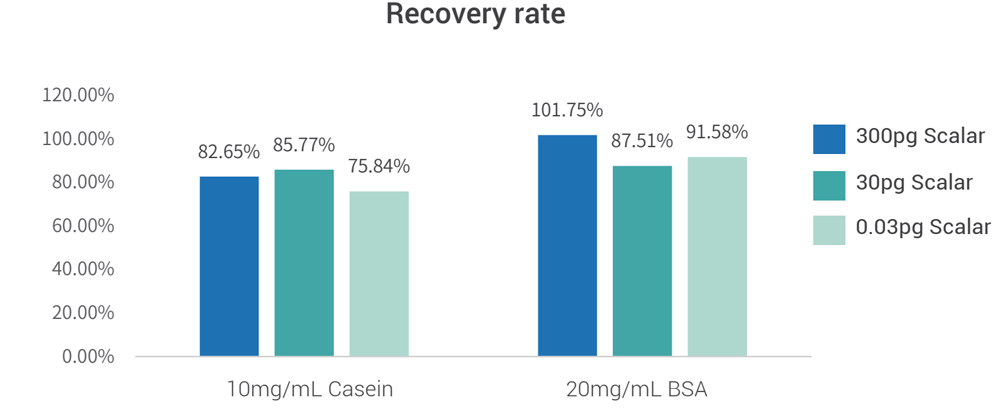
The two sample matrices (PBS+10mg/mL BSA and PBS+10mg/mL casein) were added with a total of 0.03pg, 3pg, and 300pg of CHO gDNA reference substance for pretreatment, and the final recovery of the standard addition was 70 %~130%.